Example¶
SysCorr comes with a simple demonstration example in the example/ folder. We will go through the code.
Data¶
First, gen.py generates example data for analysis:
$ python gen.py
For instance, it creates the sysline/ dataset, which contains 40 datapoints:
$ ls sysline/chain
chain00 chain04 chain08 chain12 chain16 chain20 chain24 chain28 chain32 chain36
chain01 chain05 chain09 chain13 chain17 chain21 chain25 chain29 chain33 chain37
chain02 chain06 chain10 chain14 chain18 chain22 chain26 chain30 chain34 chain38
chain03 chain07 chain11 chain15 chain19 chain23 chain27 chain31 chain35 chain39
You can look at each data point, which is made up of a chain of values to represent the uncertainty (like a markov chain):
$ gnuplot
> plot [0:1] [-1:1] "sysline/chain01"
When you prepare your own data, make sure they are scaled so that they are in the units where you expect a relationship. I.e. if you are looking for a power law, take the logarithm of the values. If your x/y values lie outside the -20:20 range, you have to adapt the transform functions in the code (or scale the data).
Run¶
Launch the example application now:
$ RESUME=1 python analyse.py out/sysline_ sysline/chain??
This will take some time, so lets find out what the analysis.py file does. You will write a similar file for your application:
output_basename = sys.argv[1]
files = sys.argv[2:]
resume = os.environ.get('RESUME', '0') == '1'
- The output will be written to ‘out/sysline_’ (first argument)
- Files are the data files (second argument and onwards).
- If you set RESUME=1, then the analysis will continue. Otherwise, it will start from scratch.
Models¶
Next, the models are defined:
example_models = dict(
# y is drawn independently of x, from a gaussian. 2 free parameters (stdev, mean)
independent_normal = PolyModel(1, rv_type = scipy.stats.norm),
# y is drawn independently of x, from a uniform dist. 2 free parameters (width, low)
independent_uniform = PolyModel(1, rv_type = scipy.stats.uniform),
# y is drawn as a function of x, with gaussian systematic scatter
# y ~ N(b*x + a, s) 3 free parameters (s, a, b)
line = PolyModel(2),
# y is drawn as a function of x, with gaussian systematic scatter
# y ~ N(c*x**2 + b*x + a, s) 4 free parameters (s, a, b, c)
square = PolyModel(3),
# two gaussians before and after 0. 4 parameters (means and stdevs)
tribinned = BinnedModel(bins = [(-0.5, 0.3), (0.3, 0.8), (0.8, 1.5)], rv_type = scipy.stats.norm)
)
If you want to compare a linear correlation to a non-correlation, you would have ‘independent_uniform’ and ‘line’.
The next code chunk just allows you to specify models from the command line without editing code:
modelnames = os.environ.get('MODELS', '').split()
if len(modelnames) == 0:
modelnames = example_models.keys()
models = []
for m in modelnames:
print 'using model:', m
model = example_models[m]
models.append((m, model))
Calculation¶
All that is left is to load the data and send it off!
print 'loading data...'
chains = [(f, numpy.loadtxt(f)) for f in files]
print 'loading data done.'
syscorr.calc_models(models=models,
chains=chains,
output_basename=output_basename,
resume=resume)
Output¶
As an example, the text output can look like this:
(...)
Total Samples: 8011
Nested Sampling ln(Z): -38.456223
ln(ev)= -38.109756655272683 +/- 0.13633196813580595
Total Likelihood Evaluations: 8011
Sampling finished. Exiting MultiNest
analysing data from out/sysline_independent_normal.txt
Analysing multinest output...
analysing data from out/sysline_independent_normal.txt
posterior plot written to "out/sysline_independent_normalpost_model_data.pdf"
calculation results: ========================================
model name | log Z
independent_uniform : -21.8
independent_normal : -16.6
tribinned : -11.3
square : -7.9
line : -6.7 <- BEST MODEL
Parameters for model "line":
syserror: 0.18+/-0.04
a: 1.8+/-0.2
b: 0.17+/-0.08
This gives you the best model (line), and the parameters of that line. (Input was y=2*x + 0.1 with error 0.1)
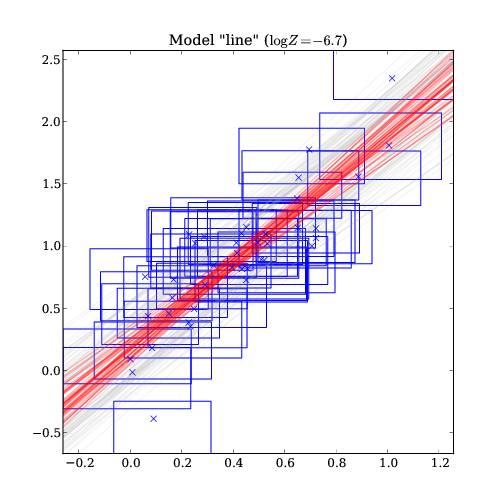
The model with several permitted parameters overplotted over the data. For each model prediction, red is the median line, while grey shows the 10%/90% quantiles. The data points are shown in blue using the x/y median and the permitted x/y extrema for the rectangles.
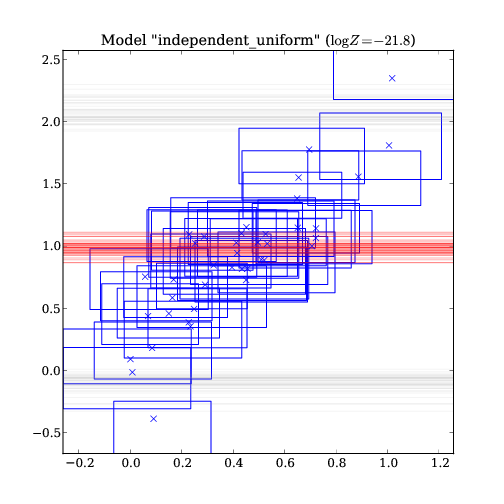
Same as above, but for the model with independent uniform distribution for y, i.e. y is uncorrelated of x. This model is ruled out by the model selection (see text output above).